5.1 Identity by descent#
Related individuals will tend to share segments of their genomes derived from a common ancestor, characterized by long stretches of identical genotypes. We will use the term identity by descent or IBD to refer to these chromosomal segments.
We are only considering autosomal chromosomes here. For those, we can consider any particular region of the genome as being shared at three possible IBD values for a pair of individuals:
IBD=0 indicates neither copy of the genome is shared in a region
IBD=1 indicates one but not both copies are shared
IBD=2 indicates both copies are shared
In the figure below, colors are used to represent stretches of the genome that share a common ancestor.

A related concept to IBD is identity by state or IBS. IBS refers to sharing of alleles at a particular SNP, regardless of whether that SNP falls on a chromosome segment that has a recent common ancestor. For example, if at a particular SNP both individuals are homozygous for the reference allele, they have IBS=2 at that SNP. Note, it is possible for a position to be shared IBS but not IBD as in the example below:
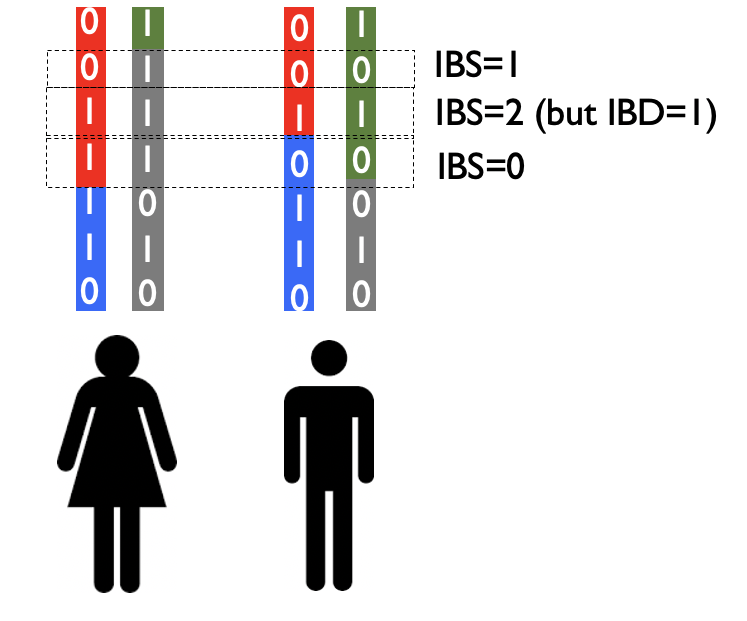
As we’ll explore in the next sections, we can predict the percent of the genome shared at IBD, as well as the length and number of segments shared, as a function of the familial relationship between a pair of individuals.
We will first discuss the percentage of the genome we expect different relative pairs to share IBD, and then will show a method (from plink) for how to estimate IBD sharing from genomes. In subsequent sections, we will look at not just the % sharing IBD, but also the length and number of shared IBD segments for different types of relative pairs.
